About nanoString Assays
The nCounter technology is highly suitable for a diverse range of discovery and translational research applications, encompassing gene expression profiling, tumor profiling, immuno-oncology profiling, single-cell gene expression analysis, miRNA expression analysis, gene fusion and copy number variation (CNV) analysis, lncRNA expression analysis, and ChIP-String expression analysis.
This versatile platform enables the simultaneous digital detection of RNA, DNA, and protein within a single experiment to provide maximum flexibility in each project.
nCounter assays streamline sample analysis by minimizing experimental variables. As a result of this optimization process, precise and accurate measurements of gene expression can be rapidly acquired for targets of interest.
NanoString Assay Advantages
- Cost-effective automated solution for multiplex analysis of up to 800 targets.
- Exceptionally precise and accurate measurements of gene expression (miRNA, RNA, CNV).
- Comprehensive and extensive target panel options available.
- Ready-to-use expertly curated gene expression panels, each containing up to 770 genes and customizable with an additional 55 unique targets.
- Enhanced flexibility: panels can be further customized by adding extra targets.
- Highly reproducible data without the need for amplification, cDNA conversion, library preparation, or technical replicates.
- Fully digital target counting providing quantitative data output.
- Streamlined sample preparation: assays can be directly performed on cell lysates or tissue homogenates, with optional RNA purification.
- Simple streamlined workflow automation with a limited number of steps, enhancing result reliability.
- Efficient data analysis yielding results in less than 24 hours.
How NanoString nCounter Works
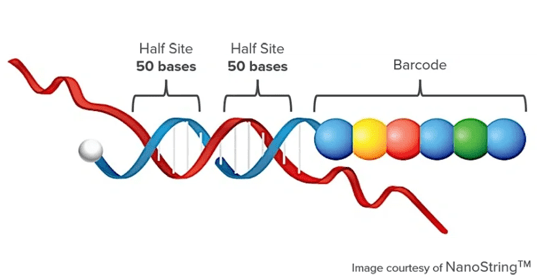
- nCounter® technology is based on the digital detection and direct molecular barcoding of individual target molecules using a unique probe pair for each specific target. This innovative technology utilizes molecular "barcodes" and single molecule imaging to accurately detect and quantify hundreds of distinct transcripts in a single reaction with exceptional sensitivity (< 1 copy per cell). The resulting digital images are processed within an nCounter instrument, and the counts of Reporter Probes are organized in a CSV file for convenient data analysis using nSolver™ Analysis Software or other preferred applications.
- Each assay comprises two 50 bp probes that specifically bind to the target: a capture probe linked to biotin for immobilization and purification purposes, as well as a reporter probe containing a distinctive fluorescent barcode. The reliability of this technology stems from its simplicity. Once the target-probe complexes have been hybridized, they are captured on a streptavidin-coated surface through the biotin tag present on the capture probe. Subsequently, these complexes are aligned, immobilized for imaging, and any excess probe is removed. A scanner then digitally quantifies each fluorescent barcode hybridized to an RNA or DNA molecule, aggregating these counts to determine the total count of targets in the sample. Therefore, one count corresponds to one molecule detected. This approach ensures an extremely low false positive rate (0.1%) compared to analog technologies that rely on measuring fluorescent intensities. The data obtained from digital reads undergo normalization against internal controls and reference genes, enabling highly precise relative counts that can be interpreted in terms of relative changes across different samples.
Our Service Features

- State-of-the-art platforms: NanoString nCounter Prep Station and Analysis System, as well as NanoString GeoMx DSP for spatial-omics.
- Extensive collection of expertly curated ready-to-use gene expression panels: Over 20,000 genes can be combined to create pathway- and disease-themed panels; custom probes can be designed for any sequence.
- Timely data delivery within 2 weeks or sooner.
- > 20 years of accumulated experience providing expert data analysis and interpretation, along with high-quality scientific and technical support.
- Optional confirmation tests available on other exceptional platforms such as Luminex 200 and FlexMAP 3D, ABI QuantStudio, CytoFlex S, Simple Western, etc.
- Sample processing services offered including protein and RNA extraction, assessment of RNA integrity and quantity (IQ, DV200).
- Library consisting of > 4000 validated antibodies enabling examination of samples at the protein level using quantitative automatic WB
- High-throughput assays available in both 96-well and 384-well formats ensuring highly reproducible data.
- Flexible service allowing quantification of any gene/protein of interest; assay layouts are custom-designed to deliver data from as few or as many samples/replicates as required.
Gene Expression Profiling
The analysis of the transcriptome serves as an initial investigative approach in exploratory studies such as drug target discovery or identification of biomarkers for specific diseases. Due to its rapid responsiveness to both external and internal stimuli, the transcriptome holds potential for predicting certain conditions. NanoString technology offers an enzyme-free, cDNA conversion and amplification-free method that requires as little as 25 ng of input material. It is compatible with various sample types including total RNA, plasma, serum, PBMC, FFPE, and cell lysate. Numerous gene expression panels comprising 200-770 genes have been developed across diverse research fields including immunology, oncology, and neurology. Most gene expression panels are available in both human and mouse versions.
MiRNA Expression Profiling
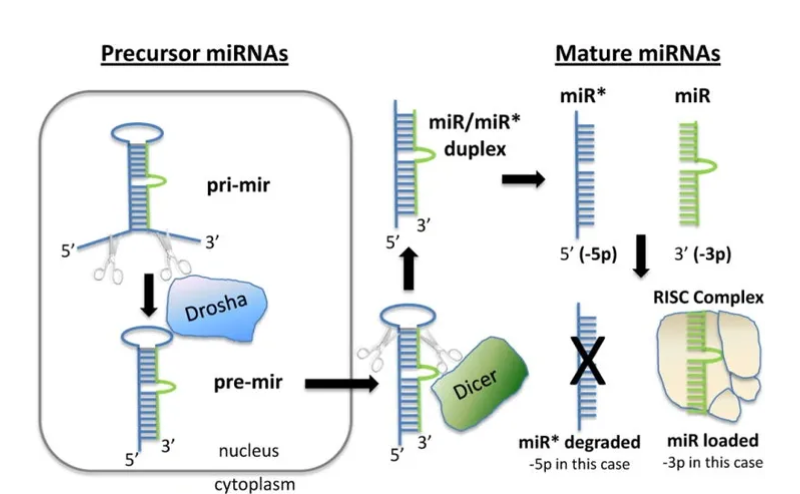
miRNA expression panels have also been developed for profiling human, mouse and rat miRNAs. PicoImmune offers sample analysis using NanoString miRNA Expression Panels which utilize specific oligos to enhance the detection of small ~22 nucleotide miRNAs. The miRNA assays include a target-specific bridge oligo and miRTag in addition to capture and reporter probes, allowing for extension of the miRNA length with the use of the bridge oligo. The resulting hybridization between the extended miRNA:miRTag molecule and two probes can then be counted as usual.
- Reliably detect and quantitate the most biologically relevant human or mouse miRNAs
- Generate results directly from FFPE, blood, or biofluids
- Utilize a chemistry that allows unambiguous discrimination of miRNAs with single nucleotide differences and six logs of dynamic range
- Streamline workflows with cost effective panels that take <1 hour hands-on time and get publication ready results within 24 hours
- Access protocols for detecting miRNA in extracellular vesicles
CNV & Gene Fusion
Aberrations in copy number are implicated in many diseases, from genetic disorders to cancer. FISH has traditionally been used to detect CNVs, but the growing number and importance of CNVs has made higher-plex technologies such as microarrays and NGS more attractive. However, these approaches require cumbersome and time-consuming workflows and a significant amount of expertise. Additionally, most microarrays are not able to resolve CNVs from FFPE samples.
NanoString has assays for Copy Number Variation (CNV) analysis and Gene Fusion Assays which are especially suited for FFPE tissue samples. PicoImmune offers sample analysis using the CNV and Gene Fusion panels from NanoString.
CNV is when a section of the genome is repeated, and where the number of repeats varies within a population. CNV can result in e.g. protein misfolding and inactivity giving rise to disease. NanoString has developed the panel Human Cancer CNV for detection of CNV of 87 genes commonly amplified or deleted in cancer. The input amount for the analysis is 300 ng genomic DNA.
Gene Fusion refers to the fusion of two separate genes by chromosomal rearrangement resulting in one hybrid gene. Gene fusions are often found to be drivers for cancer, and the resulting gene is referred to as an oncogene. The RNA input for Gene Fusion assays is app. 100 ng of total RNA, but up to 300 ng if the RNA source is FFPE tissue.
Sample Requirements: Broad Compatability
- nCounter is compatible with a wide range of sample types, including total RNA, FFPE, cell lysate, PBMC, plasma, serum, and even decades-old FFPE samples.
- It requires low input material - as little as 25 ng or 5000 cells
- Primer pools are available for multi-target enrichment in cases of low input material.
- It enables the production of high-quality data that would otherwise be challenging to obtain using methods relying on high sample input.
- Consistent results can be confidently generated for longitudinal studies using clinical-grade (often degraded) samples.
plexSet Assay: Detection of up to 96 genes in 96 samples
-
PlexSet assays accelerate cell line screening and high-throughput applications for gene expression analysis, enabling the generation of 9,216 data points across 96 samples per run.
-
Minimize hands-on time by eliminating the need for cDNA conversion, replicate utilization, or RNA purification.
-
Possess widespread applications such as cell screening, biomarker validation, drug screening, RNAi and CRISPR hit validation, and phenotypic functional testing.
-
Facilitate highly efficient and cost-effective multiplexed gene expression assays for projects with up to 96 RNA targets.
-
Utilize a simple yet robust method for multiplexing targets without the requirement to optimize probes or amplification conditions.
-
Offer expertly curated panels covering ~ 400 biological pathways and fields of interest for human, mouse, and rat samples.
-
Incorporate all necessary controls and reference genes in each panel
-
Deliver exceptional precision and reproducibility across a wide dynamic range.
-
Customize the flexible PlexSet Design by adding or omitting genes of interest.
-
Save resources with an efficient workflow: cheaper, does not require validation, and shorter turnaround time (~1/6th of qPCR)
